John Aitchison

John Aitchison
Ph.D., McMaster University
Adjunct Professor
Center for Infectious Disease Research/Institute for Systems BiologySeatte, Washington
Office: 206-256-7470 / 206-732-1344
Fax: 206-256-7229 / 206-732-1299
john.aitchison@seattlechildrens.org
Awards
- CIHR Scholar
- AHFMR Scholar
Systems Cell Biology
The human genome project revolutionized how scientists view and practice biology. For the first time, we can see the entire genetic blueprint of an organism. The difficulty now is to understand how this blueprint is read to build a living organism. This challenge has catalyzed the development of new tools for high-throughput biological measurements, such as DNA microarrays and large scale proteomic mass spectrometry. But sorting through the enormous amounts of information these techniques generate demands the integration of global analyses with computer science, mathematics, and statistics. This integration of technology, math and engineering with biology has spawned a new approach to studying biology, which is termed "systems biology". Systems cell biology is the application of systems biology at the level of the cell. It provides the basis for understanding cellular complexity, founded in molecular mechanisms that underpin biological processes. Thus the challenge for systems cell biologists is to integrate the many levels and hierarchies of cellular organization, compartmentalization, and temporal regulation into predictive models of complex cellular behaviours.
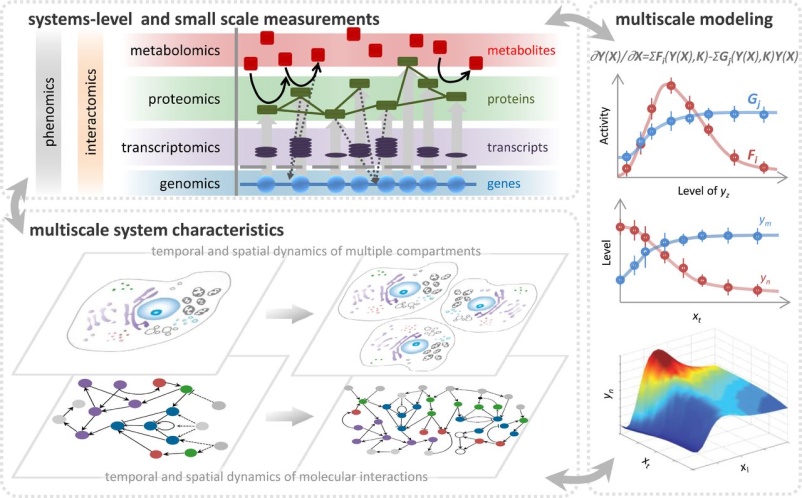 |
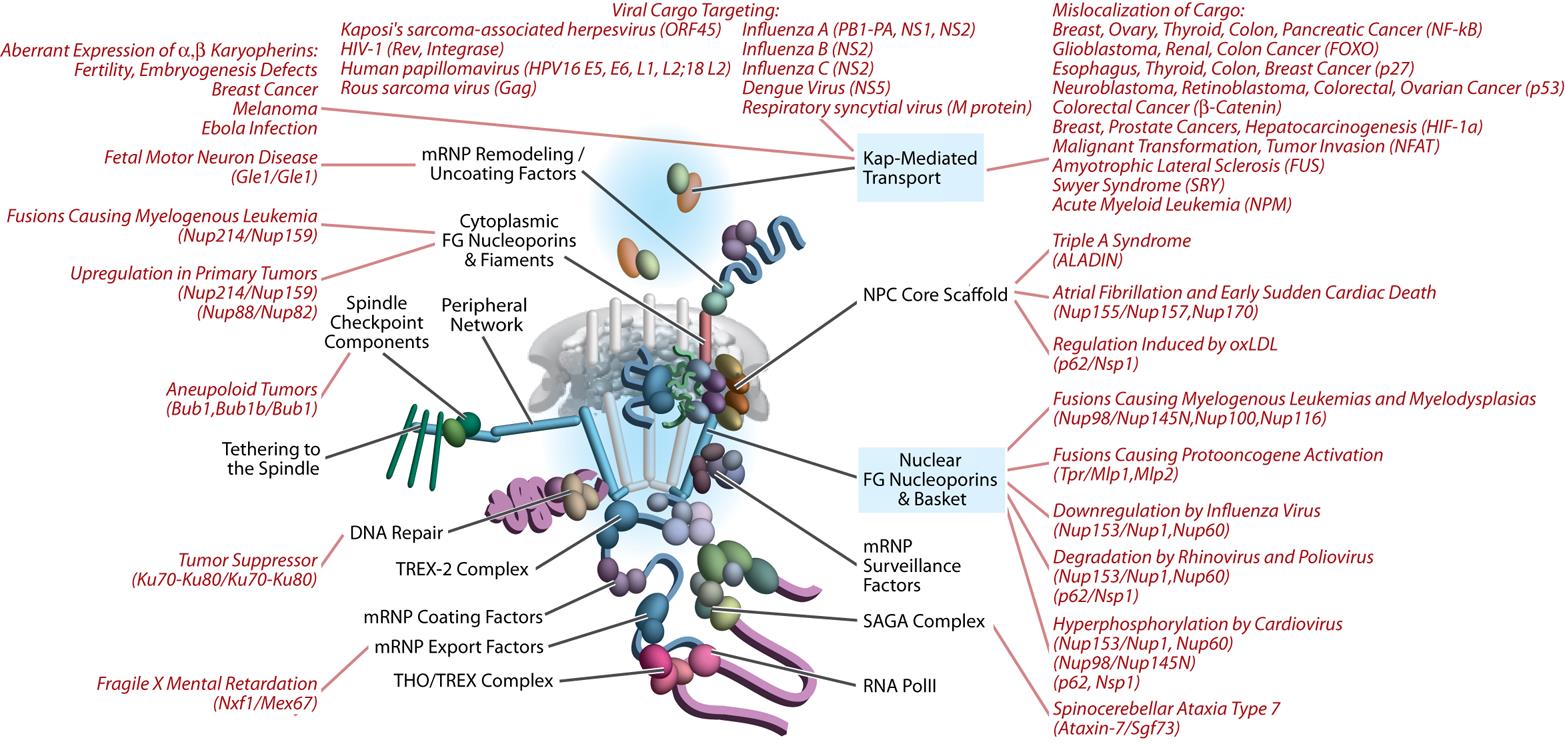
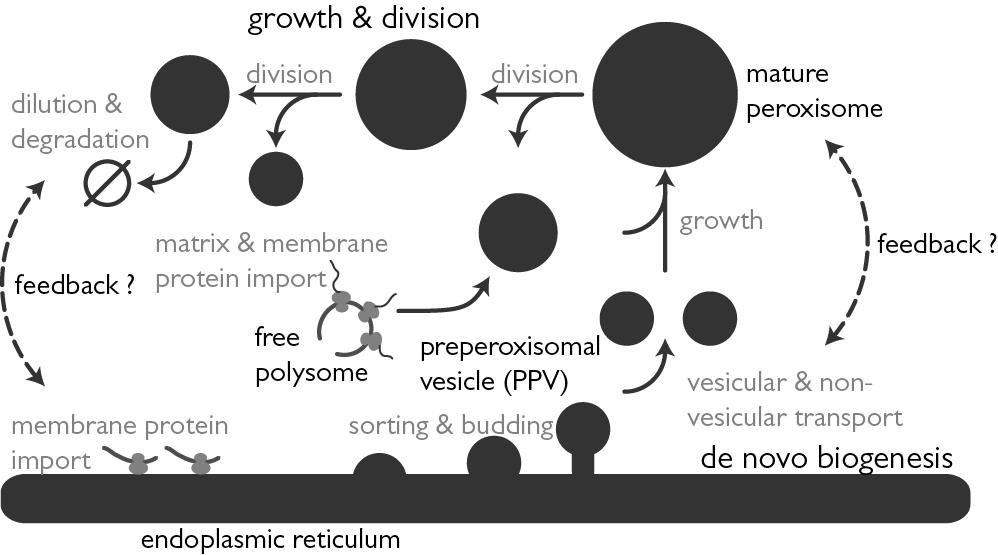
Our laboratories are located at the Center for Infectious Disease Research and the Institute for Systems Biology in Seattle WA, and we work in close collaboration with the Rachubinski and Wozniak laboratories at the University of Alberta. Together we are using systems cell biology approaches to understand how compartmentalization regulates cellular function. In particular, we are using functional genomics, proteomics, computational biology, and molecular biology to understand nucleocytoplasmic transport and peroxisome biogenesis. We also apply and develop our approaches to understand the complex interplay between pathogens, including viruses and protozoan pathogens, and their human hosts.
 |
 |
A more detailed description of our research can be found by clicking below.
 |
Selected Publications
See also:
https://www.ncbi.nlm.nih.gov/pubmed/?term=aitchison+jd
Wang Z, Danziger SA, Heavner BD, Ma S, Smith JJ, Li S, Herricks T, Simeonidis E, Baliga NS, Aitchison JD, Price ND. Combining inferred regulatory and reconstructed metabolic networks enhances phenotype prediction in yeast. PLoS Comput Biol. 2017 May17;13(5):e1005489.
Herricks T, Dilworth DJ, Mast FD, Li S, Smith JJ, Ratushny AV, Aitchison JD. One-Cell Doubling Evaluation by Living Arrays of Yeast, ODELAY! G3 (Bethesda). 2017 Jan 5;7(1):279-288.
Mast FD, Ratushny, AV, Aitchison JD. Systems Cell Biology. J Cell Biol. 2014 Sep;206(6):695-706.
Carpp LN, Rogers RS, Moritz RL, Aitchison JD. Quantitative proteomic analysis of host-virus interactions reveals a role for GBF1 in dengue infection, Mol Cell Proteomics. 2014 Nov;13(11): 2836-54.
Danziger SA, Ratushny AV, Smith JJ, Saleem RA, Wan Y, Arens CE, Armstrong AM, Sitko K, Chen WM, Chiang JH, Reiss DJ, Baliga NS, Aitchison JD. Molecular mechanisms of system responses to novel stimuli are predictable from public data. Nucleic Acids Res. 2014 Feb;42(3):1442-60.
Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013 Nov 22;14(12):803-17.
Schoggins JW, Macduff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, García-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014 Jan; 505(7485):691-5.
Van de Vosse DW, Wan Y, Lapetina1 DL, Chen W, Chiang J, Aitchison JD*, Wozniak RW*. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. April Cell. 2013 Feb;152(5):969-83.
Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, Raman A, Rust AG, Bergthaler A, Aitchison JD, Aderem A. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 2012 Oct 18;490(7420):421-5.
Ratushny AV, Saleem RA, Sitko K, Ramsey SA, Aitchison JD. Asymmetric positive feedback loops reliably control biological responses. Mol Syst Biol. 2012 Apr;8:577.
DuBois, KN, Alsford, S, Holden, JM, Buisson, J, Swiderski, M, Bart, J-M, Ratushny, AV, Wan, Y, Bastin, P, Barry, JD, Navarro, M, Horn, D, Aitchison JD, Rout, MP, Field, MC. NUP-1 is a large coiled-coil nucleoskeletal protein in trypanosomes with lamina-like functions. PLoS Biol. 2012 Mar;10(3):e1001287.
Aitchison JD, Rout MP. The Yeast Nuclear Pore Complex and Transport Through it. Genetics. 2012 Mar;190(3):855-83.
Ratushny AV, Shmulevich I, Aitchison JD. Trade-off between Responsiveness and Noise Suppression in Biomolecular System Responses to Environmental Cues. PLoS Comput Biol. 2011 Jun;7(6):e1002091.
Saleem RA, Rogers RS, Ratushny AV, Dilworth DJ, Shannon PT, Shteynberg D, Wan Y, Moritz RL, Nesvizhskii AI, Rachubinski RA, Aitchison JD. (2010) Integrated phosphoproteomic analysis of a signaling network governing nutrient response and peroxisome induction, Mol Cell Proteomics. 2010 Apr 15. Sekedat MD, Fenyö D, Rogers RS, Tackett AJ, Aitchison JD, Chait BT. (2010) GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome, Mol Syst Biol. 2010;6:353. Epub 2010 Mar 9. PMCID: PMC2858444
Shmulevich, I. and Aitchison, J. D. (2009) Deterministic and stochastic models of genetic regulatory networks, Methods Enzymol, 467: 335-356
Wan, Y., Chiang, J. H., Lin, C. H., Arens, C. E., Saleem, R. A., Smith, J. J. and Aitchison, J. D. (2009) Histone chaperone Chz1p regulates H2B ubiquitination and subtelomeric anti-silencing, Nucleic Acids Res, 2010 Mar 1;38(5):1431-40. Epub 2009 Dec 14.. PMCID: PMC2836552
Smith, J. J. and Aitchison, J. D. (2009) Regulation of peroxisome dynamics, Curr Opin Cell Biol. Feb;21(1):119-26. Epub 2009 Jan 31.
Fagarasanu, A., Mast, F. D., Knoblach, B., Jin, Y., Brunner, M. J., Logan, M. R., Glover, J. N., Eitzen, G. A., Aitchison, J. D., Weisman, L. S. and Rachubinski, R. A. (2009) Myosin-driven peroxisome partitioning in S. cerevisiae, J Cell Biol, 186: 541-554. PMC2733749
Ptak, C., Anderson, A. M., Scott, R. J., Van de Vosse, D., Rogers, R. S., Sydorskyy, Y., Aitchison, J. D. and Wozniak, R. W. (2009) A role for the karyopherin Kap123p in microtubule stability, Traffic, 10: 1619-1634
Ratushny, A. V., Ramsey, S. A., Roda, O., Wan, Y., Smith, J. J. and Aitchison, J. D. (2008) Control of transcriptional variability by overlapping feed-forward regulatory motifs, Biophys J, 95: 3715-3723
Saleem, R. A., Knoblach, B., Mast, F. D., Smith, J. J., Boyle, J., Dobson, C. M., Long-O'Donnell, R., Rachubinski, R. A. and Aitchison, J. D. (2008) Genome-wide analysis of signaling networks regulating fatty acid-induced gene expression and organelle biogenesis, J Cell Biol, 181: 281-292
Smith, J. J., Ramsey, S. A., Marelli, M., Marzolf, B., Hwang, D., Saleem, R. A., Rachubinski, R. A. and Aitchison JD. Transcriptional responses to fatty acid are coordinated by combinatorial control, Mol Syst Biol, 2007 3: 115.
Ramsey, S. A., Smith, J. J., Orrell, D., Marelli, M., Petersen, T. W., de Atauri, P., Bolouri, H. and Aitchison JD. (2006) Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast, Nat Genet, 38: 1082-1087.
Dilworth, D. J., Tackett, A. J., Rogers, R. S., Yi, E. C., Christmas, R. H., Smith, J. J., Siegel, A. F., Chait, B. T., Wozniak, R. W. and Aitchison JD. (2005) The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol, 171: 955-965.
Marelli, M., Smith, J. J., Jung, S., Yi, E., Nesvizhskii, A. I., Christmas, R. H., Saleem, R. A., Tam, Y. Y., Fagarasanu, A., Goodlett, D. R., Aebersold, R., Rachubinski, R. A. and Aitchison JD. (2004) Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J Cell Biol, 167: 1099-1112.
Laboratory Members
Postdoctoral Fellows
Sanjeev Kumar
Fred Mast
Research Scientists
Song Li
Leslie Miller
Bioinformatics Scientist
Max Neal
Senior Scientist
Paul Olivier
Staff Scientists
Fergal Duffy
Thurston Herricks
Atri Navare
Research Fellow
Christoph Carter
